Mapping Out How Viruses Jump Between Species
7:22 minutes
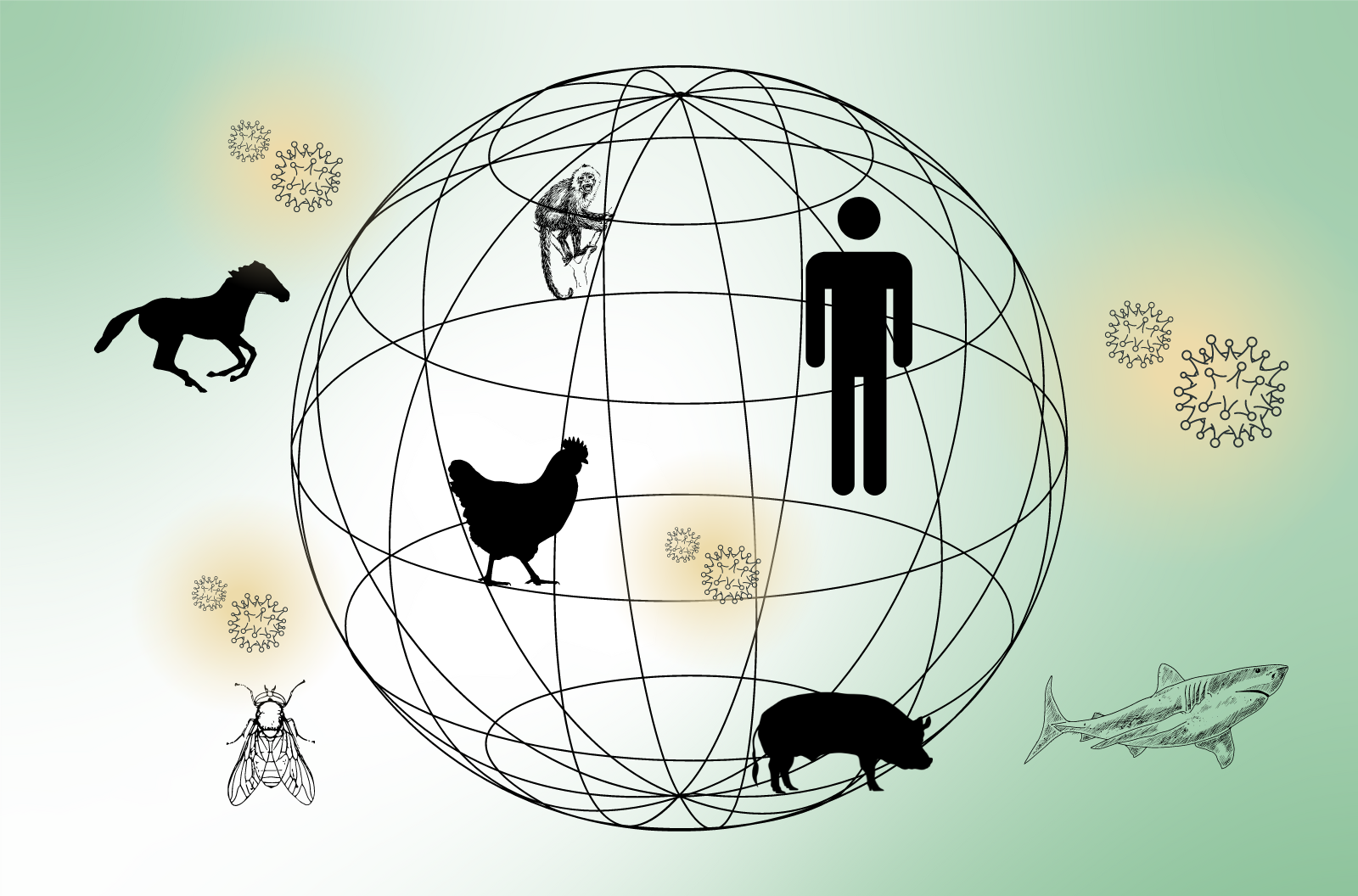
In the world of emerging infectious diseases, one of the looming threats comes from the so-called zoonotic diseases—pathogens that somehow make the jump from an animal host to a human one. This includes pathogens such as COVID-19 and avian influenza, a.k.a. bird flu, which can sometimes cross the species divide. But a new analysis published in the journal Nature Ecology and Evolution finds that when it comes to viruses, more viral species appear to have jumped from humans to animals than the other way around. And even more cases of interspecies transmission don’t involve humans at all.
Cedric Tan, a PhD student in the University College London Genetics Institute and Francis Crick Institute, joins guest host Arielle Duhaime-Ross to talk about the analysis, and what it tells us about our place in a global web of viruses.
Invest in quality science journalism by making a donation to Science Friday.
Cedric Tan is a PhD student in the University College London Genetics Institute and Francis Crick Institute in London, UK.
ARIELLE DUHAIME-ROSS: This is Science Friday. I’m Arielle Duhaime-Ross, sitting in for Ira Flatow. When we talk about emerging diseases, one of the looming threats is the so-called zoonotic disease, pathogens that somehow make the jump from an animal host to a human one. Think COVID 19 or avian influenza, a.k.a. Bird flu, which sometimes crosses the species divide.
But a new analysis published in the journal Nature, Ecology, and Evolution finds that when it comes to viruses, more of them appear to have jumped from humans to animals instead of the other way around. Joining me now to talk about that finding is Cedric Tan. He’s a PhD student in the University College London Genetics Institute and Francis Crick Institute, and he’s the lead author on that report I just mentioned. Welcome to Science Friday.
CEDRIC TAN: Well, thanks, Arielle, for having me.
ARIELLE DUHAIME-ROSS: Yeah, great to have you. OK, so to start out, how do you track the path of a virus from one species to another?
CEDRIC TAN: Right. So to do this, we use statistical methods, based on the genomic sequences of the virus. And what we can do is, using these methods, we can try to reconstruct where the virus has come from, say, as if it has infected birds at some point and then was passed on into, say, humans. And this is what we call ancestral reconstruction. And so using these tree of lives generated from viral phylogenies, we can try to infer the direction of flow of viruses.
ARIELLE DUHAIME-ROSS: So what I’m getting from you is you can look at the viral genome of a virus that infects humans and then also the genome of a virus that infects pigs or bats, and these genomes are very similar. It’s the same virus. And by looking at the different changes you can sort of reconstruct what happened and how those jumps occurred.
CEDRIC TAN: Yes, we know what is an ancestor virus and what we know what is a descendant virus. And based on this, we can say, OK, the ancestor has been found in pigs, for example, and the descendant has been found in humans, for example. And so based on that, we can say that this virus has actually jumped from pigs into humans.
ARIELLE DUHAIME-ROSS: OK. So in this project, you were looking at thousands of viral genomes. How many did you study?
CEDRIC TAN: There are two different analyses presented in this paper. So the first one was to assess the state of genomic surveillance around the world and so basically, what we have collected so far, what we have sequenced so far. And so for this analysis, we used about 12 million viral sequences. And this was uploaded by various labs around the world to a public database called NCBI Virus.
And then we further subsampled this set of viral sequences to around 60,000 sequences. So these are 60,000 high-quality, complete viral genomes. And these were used for our detailed analysis of viral host jumps.
ARIELLE DUHAIME-ROSS: And so this library of viral genomes, how does that compare to the whole universe of viruses out there? How big is this library?
CEDRIC TAN: Yeah, this is an excellent question. And the short answer is we don’t really know. Because we’ve largely focused on the viruses that infect humans, so we don’t really know the true number of viruses that infect animals beyond humans. So in fact, only 9% of fish families are represented in the database, and 13% of amphibians were represented. We are still discovering new viruses every day, so the estimate of the true viral diversity that we expect keeps changing as we discover that.
ARIELLE DUHAIME-ROSS: Did certain types of viruses seem more likely to cross to another host than others?
CEDRIC TAN: Yes. So one of the other messages in our paper is that viruses that can already infect a broad range of hosts are more at risk of jumping into a new host. In fact, we’ve known that many viruses that have recently jumped into humans have remarkably broad host ranges. One example of this is SARS-CoV-2, which caused the COVID-19 pandemic. And this virus can infect nearly every mammalian order while requiring minimal adaptations. So what we found in this study is really that these broad host range viruses are what we should be focusing on to potentially identify the next pandemic virus because they are more likely to jump into a new host species.
ARIELLE DUHAIME-ROSS: When you say more likely to jump, what does that actually mean? What does a virus need to have– what characteristics does it need to have to be able to make a cross-species shift? Is there a special recipe, a magic sauce?
CEDRIC TAN: Well, it’s not really a magic sauce, but first of all, a virus in an infected host must first be able to transmit the virus, either via infected fluids or being excreted through feces or through various other modes of transmission. And then the virus must then be able to enter the cells of the new host. And the virus must also then be able to survive and replicate in the new host cell. So these are some of the kind of barriers to entry when a virus is trying to exploit a new host species.
One other thing is that most viruses don’t really have their own replication machinery. And so they must hijack the proteins in the new host cell to be able to replicate themselves. And after replication, the virus must then be able to transmit from the new host to other hosts of the same species sufficiently well so that they can circulate in this new host population.
ARIELLE DUHAIME-ROSS: You estimate the number of shifts, animal to human, human to animal, and animal-animal jumps. What’s the magnitude of those kinds of shifts?
CEDRIC TAN: So in our study, we inferred about 2,900 host jumps in total. 79% of these jumps involves only animals, and only 21% involves humans. So that’s about 600 host jumps. Of these 600 host jumps that involve humans, 64% were humans to animal. And half of that, which is 36%, involve animal to human transmission events.
ARIELLE DUHAIME-ROSS: What does this all tell you? What are the big take-home messages?
CEDRIC TAN: First thing is that we have a very huge impact on the environment around us. Second thing is that we can start to think of leveraging the huge amounts of genomic data that have been deposited on public databases so far, to try to understand the wider flow of viruses around the globe and between different animals. And I think that’s really important because we’ve been focusing on zoonosis. We’ve been focusing on things that infect us. But we are just a single species in this really broad and diverse network of hosts around the world. And we have to stop being so myopic and try to understand this wider ecological network to better understand and improve our capabilities to predict the next emerging infectious disease.
ARIELLE DUHAIME-ROSS: Cedric Tan is a PhD student in the University College London Genetics Institute and Francis Crick Institute. Cedric, thank you so much for talking with me today.
CEDRIC TAN: Well, thank you, Arielle. It was a very nice chat.
Copyright © 2024 Science Friday Initiative. All rights reserved. Science Friday transcripts are produced on a tight deadline by 3Play Media. Fidelity to the original aired/published audio or video file might vary, and text might be updated or amended in the future. For the authoritative record of Science Friday’s programming, please visit the original aired/published recording. For terms of use and more information, visit our policies pages at http://www.sciencefriday.com/about/policies/
As Science Friday’s director and senior producer, Charles Bergquist channels the chaos of a live production studio into something sounding like a radio program. Favorite topics include planetary sciences, chemistry, materials, and shiny things with blinking lights.
Arielle Duhaime-Ross is freelance science journalist, artist, podcast, and TV host based in Portland, OR.